Biology 4289b
Finding information for your project
Finding sequences and species of interest
1- If you have identified a group of organisms you like, find the Latin name of one of the lesser known species.
2- Go to the NCBI Taxonomy browser.
3- Search the Latin name and click on the desired species.
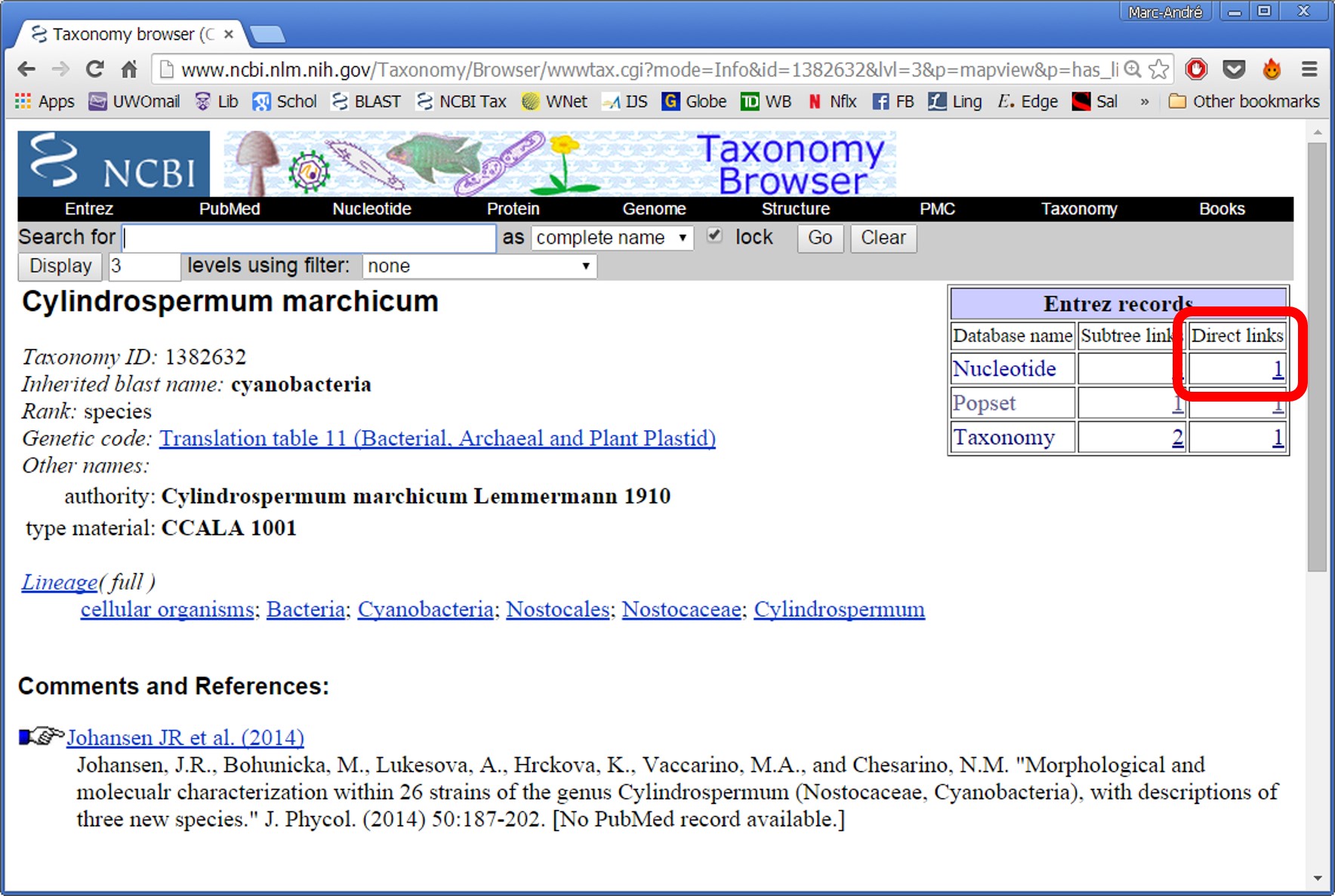
4- Click on the link in the top right table, entitled "Nucleotide - Direct Links"
5- This will take you to all available sequences for that species.
6- Select a suitable sequence and BLAST it (top right: Analyse this sequence - run BLAST).
7- Select the correct database and BLAST.
8- Select (check mark) the sequences that you wish to download.
9- Click GenBank
10- Click Display settings.
11- Click FASTA (text).
12- Select all sequences (Ctrl-a) and place them on the clipboard (CTRL-c)
13- Open Mega6
14- Align - Edit/Build Alignment - Create a new alignment - DNA (as appropriate).
15- Click into the editor and paste your sequences (Ctrl-v).
16- Click on a single nucleotide.
17- Click on the Muscle icon (flexed arm).
18- Align DNA - OK - Compute.
19- Examine your sequences for a phylogenetic signal.
20- Trim the fringe.
21- Save the sequences.
22- Search for information, using:
1- If you have identified a group of organisms you like, find the Latin name of one of the lesser known species.
2- Go to the NCBI Taxonomy browser.
3- Search the Latin name and click on the desired species.

4- Click on the link in the top right table, entitled "Nucleotide - Direct Links"
5- This will take you to all available sequences for that species.
6- Select a suitable sequence and BLAST it (top right: Analyse this sequence - run BLAST).
7- Select the correct database and BLAST.
8- Select (check mark) the sequences that you wish to download.
9- Click GenBank
10- Click Display settings.
11- Click FASTA (text).
12- Select all sequences (Ctrl-a) and place them on the clipboard (CTRL-c)
13- Open Mega6
14- Align - Edit/Build Alignment - Create a new alignment - DNA (as appropriate).
15- Click into the editor and paste your sequences (Ctrl-v).
16- Click on a single nucleotide.
17- Click on the Muscle icon (flexed arm).
18- Align DNA - OK - Compute.
19- Examine your sequences for a phylogenetic signal.
20- Trim the fringe.
21- Save the sequences.
22- Search for information, using:
- Google images
- Google Scholar
- Wikipedia
- The sequence page of a deposit (step 6, above), usually contains a link to the publication relevant to this sequence (PUBMED entry). This may be very useful.