![]()
Expressing the human genome
URL: http://www.nature.com/cgi-taf/DynaPage.taf?file=/nature/journal/v409/n6822/full/409832a0_fs.html
Date accessed: 15 March 2001
| Nature 409, 832 - 833 (2001) © Macmillan Publishers Ltd. |
|
|
ROSSELLA TUPLER*†, GIOVANNI PERINI‡ & MICHAEL R. GREEN*
We have searched the human genome for genes encoding new proteins that may be involved in three nuclear gene expression processes: transcription, pre-messenger RNA splicing and polyadenylation. A plethora of potential new factors are implicated by sequence in nuclear gene expression, revealing a substantial but selective increase in complexity compared with Drosophila melanogaster and Caenorhabditis elegans. Although the raw genomic information has limitations, its availability offers new experimental approaches for studying gene expression.
The availability of the human and other genome sequences will revolutionize all fields of biomedical research. But, as the genome is itself the object of gene expression, the impact may be particularly profound for those of us studying this process. Here we preview what the human genome has to offer to those interested in gene expression.
Gene expression comprises various events from the initial transcription of a gene in the nucleus to the translation of mRNA in the cytoplasm. Here we focus on three steps that occur in the nucleus: transcription, pre-mRNA splicing and 3' end formation. All three involve the recognition of a nucleic acid (DNA or RNA) that serves as a scaffold for a multiprotein complex in which the relevant reaction (transcription, splicing or 3' end formation) occurs. Research in each is aimed at identifying all the relevant components and elucidating how the reaction is controlled.
General transcription factors
Factors involved in the transcription of eukaryotic protein-coding genes by RNA
polymerase II fall into two groups: general (or basic) transcription factors (GTFs)
and transcriptional activators. GTFs are required for accurate transcription
initiation in vitro1. They include RNA
polymerase II itself and at least six GTFs: TFIID, TFIIA, TFIIB, TFIIE, TFIIF
and TFIIH. The GTFs assemble on the promoter to form a preinitiation complex (PIC).
Of the GTFs, TFIID is the primary sequence-specific DNA-binding component; it
initiates PIC assembly by interaction with the TATA box. TFIID is composed of
the TATA-box-binding protein (TBP) and multiple TBP-associated factors (TAFs)2.
This basic transcription machinery has, in general, been highly conserved from
yeast to human.
We have searched the human genome sequence for GTFs. Consistent with the Drosophila, C. elegans and Saccharomyces cerevisiae genomes3, the human genome contains single-copy genes encoding the components of RNA polymerase II, TFIIB, TFIIE, TFIIF and TFIIH, in general without evidence for related genes (see Supplementary Information Table 1). A potentially important exception is the presence of three genes related to cdk7, a cyclin-dependent kinase that is associated with TFIIH4.
We found gene sequences related to many of the TFIID subunits, including TBP, TFIIA and several TAFs, indicating that the potential diversity of human TFIID is much greater than that of Drosophila3 (see Supplementary Information Table 1). For example, we identified six human genes related to TAF32, but no Drosophila genes related to the homologous TAF40. It was thought that all metazoans possess a single gene for TBP, with a second gene encoding a TBP-like factor (TLF). Unusually, Drosophila contains a third TBP-related gene (TRF1)5. However, our searches reveal that humans also have a third TBP-related sequence on chromosome 14 (see Supplementary Information Fig. 1).
Transcriptional activators
Transcriptional activity is strongly stimulated by promoter-specific activators.
In general, these are sequence-specific DNA-binding proteins whose recognition
sites are present in target promoters. Activators have been classified into
families on the basis of their DNA-binding domains. A search of the human genome
sequence revealed more than 2,000 hypothetical genes that encode transcriptional
activators (Fig.
1). The C2H2 zinc finger proteins form the largest family (around 900
members), and this is also the largest family of activators in Drosophila,
C. elegans and S. cerevisiae. There are around twice as many
basic region leucine zipper (bZIP) proteins, nuclear receptors and
helix–loop–helix proteins in the human genome as in the Drosophila
genome, and 5–10 times more than in the C. elegans and S. cerevisiae
genomes (Fig.
1).
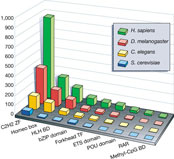 |
Figure 1
Genome-wide comparison of transcriptional activator families in
eukaryotes. Full legend High resolution image and legend (74k) |
We performed more extensive searches on the bZIP protein superfamily of transcriptional activators. We have aligned these new bZIP proteins on the basis of sequence relatedness to existing members of the bZIP subfamilies, such as Jun, Fos, ATF, CREB and c/EBP (see Supplementary Information Fig. 2). This revealed 18 new bZIP genes, primarily belonging to the Fos and CREB families. Three new genes belong to the small TEF/HLF family, which shares strong similarity with the C. elegans gene ces-2, which is involved in the control of programmed cell death6, 7. This search illustrates how the human genome sequence will provide many new factors that may be involved in gene expression, although their roles remain unknown.
Pre-mRNA splicing
Pre-mRNA splicing occurs in a large, dynamic complex, the spliceosome. To
assemble the spliceosome, four small nuclear ribonucleoprotein (snRNP) particles
(U1, U2, U5 and U4/U6) and many non-snRNP proteins interact with pre-mRNA in an
ordered pathway8, 9. In
metazoans, spliceosome assembly begins with recognition of the 5' splice site by
U1 snRNP, and of the polypyrimidine (Py)-tract by the U2 snRNP auxiliary factor,
U2AF10.
We have searched the human genome sequence for several protein-splicing factors that act early during spliceosome assembly. The results reveal significantly greater complexity than is found in Drosophila11 (see Supplementary Information Table 2). Of particular interest are several new genes closely related to the small subunit of U2AF, U2AF35 (see Supplementary Information Fig. 3).
Polyadenylation
Eukaryotic mRNAs have a 3' poly(A) tail, around 200 nucleotides long, which is
added post-transcriptionally following endonucleolytic cleavage of the pre-mRNA.
Addition of poly(A) is directed by a polyadenylation sequence, AAUAAA,
located 10–30 bases upstream of the polyadenylation site. Polyadenylation
requires various protein components, including a cleavage/polyadenylation
specificity factor (CPSF), a cleavage stimulatory factor (CstF), other cleavage
factors and a poly(A) polymerase (PAP)12. Several
poly(A)-binding proteins (PABs) bind to the mature poly(A) tail.
We identified genes related to factors involved in mRNA 3' end formation, including PAP and PAB (see Supplementary Information Table 3). It was known that there were multiple, alternatively spliced variants of PAP, but the existence of several PAP-related gene sequences was unexpected and increases the potential diversity of this enzyme. These results suggest that the polyadenylation machinery of humans is more complex than that of Drosophila.
Limitations of the genomic information
Although these searches highlight the power of the new genomic information, they
also reveal important limitations. In particular, the existence of a related
gene sequence does not mean that there is a corresponding protein: the sequence
could be a non-expressed pseudogene. Indeed, some new gene sequences contain
stop codons or lack introns. We know that some of the related gene sequences are
expressed, however, as they are present in expressed sequence tag (EST)
databases. Even if a related sequence is an expressed gene, we do not know
whether the two related genes are simultaneously expressed in the same cell, or
are differentially expressed—for example, in a tissue- or development-specific
manner. Expression studies will be required to complement genomic information. A
final caveat is that many of the factors are components of multi-subunit
complexes. Sometimes the same factor is present in multiple complexes whose
activities differ substantially. Thus, the full value of the genomic information
can be realized only when it is coupled with appropriate biochemical studies.
New approaches for studying gene expression
Full eukaryotic genome sequences will allow new experimental strategies to study
gene expression. In the 'classical' pre-genomic strategy, factors involved in
gene expression were identified through biochemical or genetic experiments that
focused on a specific gene expression process (for example, a transcription
pathway). The human genome sequence contains many new factors whose sequences
suggest roles in gene expression but whose precise activities and functions are
unknown. Therefore, a 'post-genomic' approach can start with the new gene (or
its protein product) and determine its activity and the pathway in which it
participates.
Consider the new bZIP proteins. Clues to the biological processes in which they participate may be obtained by determining their tissue expression patterns, elucidating their DNA binding specificities13 or mapping their chromosomal sites of occupancy14. Ultimately, understanding the function of a transcriptional activator will require identification of the genes that it controls. This requires derivation of appropriate cell lines or animal models in which the protein can be expressed (or inactivated) in a regulated fashion. The consequences of its expression (or inactivation) on the transcription of specific genes is then monitored. In the most powerful version of this approach, target gene transcription can be assayed in parallel using high-density DNA microarrays15.
By performing focused searches of the draft human genome sequence, we have identified many new factors that may be involved in transcription, splicing and mRNA 3' end formation. The increased complexity of the human gene expression machinery implies that gene expression may be particularly important in shaping human development and physiology.
Supplementary information is available on Nature's World-Wide Web site (http://www.nature.com) or as paper copy from the London editorial office of Nature.
| 1. | Orphanides, G., Lagrange, T. & Reinberg, D. The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657-2683 (1996). | PubMed | |
| 2. | Burley, S. K. & Roeder, R. G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769-799 (1996). | PubMed | |
| 3. | Aoyagi, N. & Wassarman, D. A. Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation. J. Cell. Biol. 150, F45-F49 (2000). | PubMed | |
| 4. | Coin, F. & Egly, J. M. Ten years of TFIIH. Cold Spring Harb. Symp. Quant. Biol. 63, 105-110 (1998). | PubMed | |
| 5. | Berk, A. J. TBP-like factors come into focus. Cell 103, 5-8 (2000). | PubMed | |
| 6. | Inaba, T. et al.Reversal of apoptosis by the leukaemia-associated E2A-HLF chimaeric transcription factor. Nature 382, 541-544 (1996). | PubMed | |
| 7. | Metzstein, M. M., Hengartner, M. O., Tsung, N., Ellis, R. E. & Horvitz, H. R. Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2. Nature 382, 545-547 (1996). | PubMed | |
| 8. | Burge, C. B., Tuschl, T. H. & Sharp, P. A. in The RNA World 2nd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, J. R.) 525-560 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1999). |
| 9. | Staley, J. P. & Guthrie, C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315-326 (1998). | PubMed | |
| 10. | Reed, R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 6, 215-220 (1996). | PubMed | |
| 11. | Mount, S. M. & Salz, H. K. Pre-messenger RNA processing factor in the Drosophila genome. J. Cell. Biol. 150, F37-F43 (2000). | PubMed | |
| 12. | Colgan, D. F. & Manley, J. L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755-2766 (1997). | PubMed | |
| 13. | Ouellette, M. M. & Wright, W. E. Use of reiterative selection for defining protein-nucleic acid interactions. Curr. Opin. Biotechnol. 6, 65-72 (1995). | PubMed | |
| 14. | Orlando, V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromating immunoprecipitation. Trends Biochem. Sci. 25, 99-104 (2000). | Article | PubMed | |
| 15. | Young, R. A. Biomedical discovery with DNA arrays. Cell 102, 9-5 (2000). | PubMed | |
Acknowledgements. M.R.G. is an investigator of the Howard Hughes Medical Institute. This work was supported in part by a grant from NIH to M.R.G.
Category: 32. Genome Project and Genomics