![]()
Keeping time with the human genome
URL: http://www.nature.com/cgi-taf/DynaPage.taf?file=/nature/journal/v409/n6822/full/409829a0_fs.html
Date accessed: 15 March 2001
| Nature 409, 829 - 831 (2001) © Macmillan Publishers Ltd. |
|
|
JONATHAN D. CLAYTON*, CHARALAMBOS P. KYRIACOU* & STEVEN M. REPPERT†
The cloning and characterization of 'clock gene' families has advanced our understanding of the molecular control of the mammalian circadian clock. We have analysed the human genome for additional relatives, and identified new candidate genes that may expand our knowledge of the molecular workings of the circadian clock. This knowledge could lead to the development of therapies for treating jet lag and sleep disorders, and add to our understanding of the genetic contribution of clock gene alterations to sleep and neuropsychiatric disorders. The human genome will also aid in the identification of output genes that ultimately control circadian behaviours.Completion of the draft sequence of the human genome is an epic event, providing a treasure-trove of new genes for all aspects of biological investigation. For circadian clock research, 'snapshot' analysis of the draft sequence provides a glimpse of potential new members of canonical 'clock gene' families. Such candidate genes will add to the riches of the post-genomic era, ultimately leading to a wider understanding of the molecular control of the circadian clock.
The master circadian clock in mammals, in the suprachiasmatic nuclei (SCN) of the anterior hypothalamus, drives daily variations in many physiological and behavioural processes, such as the sleep–wake rhythm and daily variations in body temperature, hormone levels, cognition and memory1. Dawn and dusk coordinate or entrain the circadian clock through neural pathways connecting the retina to the SCN, so that the master clock and its output rhythms do not drift from 24 h, but remain aligned with the solar day. Transient disruption of circadian timing following transmeridian flights leads to jet lag, and chronic alterations of the central clock mechanism in shift workers (around 25% of the working population) may contribute to poor health and sleep disorders. Finally, specific rhythm defects may be involved in neuropsychiatric illnesses.
The fruitfly Drosophila melanogaster has long been the animal model of choice for genetic analysis of circadian rhythms. In recent years, with the help of forward and reverse genetics, homology-based approaches and protein-interaction screens, homologues of most of the genes involved in the fly clock have been cloned in mammals2 (Fig. 1, Table 1). Although there are compelling similarities between the general clock mechanisms of Drosophila and mice, gene duplication has led to reassignment of specific functions among structurally homologous components.
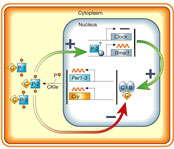 |
Figure 1
Model of circadian clockwork within the SCN. Full legend High resolution image and legend (65k) |
Analysis of the fly circadian clock appears to be close to genetic saturation, indicating that most of the cardinal genes may have been identified3. It is less clear whether the whole complement of clock genes has been identified in mammals, because expansion of gene families makes it likely that additional family members may await discovery. We searched the draft sequence of the human genome for new clock genes (see Supplementary Information for Methods).
A few words of caution are in order concerning the sequence analysis. Although we have identified new clock gene candidates, we have no information regarding their expression patterns or function. We detected all of the previously identified clock loci in mammals, confirming the near completeness of the human genome sequence data. This does not rule out the possibility, however, of further clock candidates surfacing from advanced searches of the finished genome sequence.
Genes involved in the core clock mechanism
The core clock mechanism of mammals is composed of interacting positive and
negative transcriptional/translational feedback loops4
(Fig.
1). The pivotal components of this mechanism are two basic
helix–loop–helix (bHLH)/PAS-containing transcription factors, CLOCK and
BMAL1, which are assumed to pair through their protein-interactive PAS domains.
PAS domains are found in a diverse family of proteins (PER, ARNT and SIM are the
founder members) and are a common feature of transcription factors that are
essential for the clock machinery of fungi, insects and mammals5.
CLOCK–BMAL1 heterodimers drive the rhythmic transcription of three Period genes (mPer1–mPer3 in the mouse) and two Cryptochrome genes (mCry1 and mCry2). As the mPER and mCRY proteins are translated, they form PER–CRY complexes that are translocated to the nucleus. There the mCRY proteins act as negative regulators by directly interacting with CLOCK or BMAL1, or both, to inhibit transcription, forming a negative feedback loop. At the same time, mPER2 contributes to the transcription of Bmal1, which is rhythmically expressed with a peak phase opposite to that of mPer/mCry, forming a positive feedback loop. The push–pull action of the positive and negative feedback loops perpetuates the self-sustaining nature of the circadian clock.
Analysis of the draft sequence identified human hPER1, hPER2, hPER3, hCRY1 and hCRY2. Moreover, an additional single-copy gene was identified on chromosome 7 with high sequence similarity to the Per family (Table 1). A fragment of genomic sequence is available, but no expressed sequence tag (EST) matches were obtained. Nevertheless, the putative translation of this fragment encodes 231 residues that encompass most of the PAS region and can be compared with those of other family members (see Supplementary Information Figs A and B). If 'hPER4' can be confirmed as an expressed sequence rather than a pseudogene, the human genome project will have yielded a potentially exciting finding, because such a fourth PER will contribute to our understanding of the core clock mechanism. Even if it is not expressed, identification of an hPER4 locus may help us to decipher the evolution of the Per gene family.
Our analysis identified hBMAL1 (hMOP3) and hCLOCK, as well as two previously described members of this transcription factor family, hMOP4 and hMOP9 (also called CLIF)6-8, which encode proteins closely related to hCLOCK and hBMAL1, respectively (Table 1; see Supplementary Information Figs C and D). We did not find any new genes related to these transcription factors. Because MOP4–BMAL1 and CLOCK–MOP9/CLIF heterodimers can activate transcription from E-boxes (CACGTG) in vitro, MOP4 and MOP9/CLIF have the potential to contribute to transcriptional regulation within the core clock mechanism. MOP9/CLIF is in fact expressed in the SCN7, 8.
Kinases important for clock function
Phosphorylation and proteolysis of clock proteins can determine their cellular
location and stability, and are key ingredients for building time delays into
the 24-h molecular mechanism3. In Drosophila,
DOUBLETIME, which is closely related to mammalian casein kinase I epsilon (CKI![]() ),
phosphorylates PER, thereby influencing PER turnover3.
Studies of the tau mutation in Syrian hamsters (a spontaneous,
semidominant mutation leading to marked shortening of the circadian period)
reveal that it encodes a missense mutation within CKI
),
phosphorylates PER, thereby influencing PER turnover3.
Studies of the tau mutation in Syrian hamsters (a spontaneous,
semidominant mutation leading to marked shortening of the circadian period)
reveal that it encodes a missense mutation within CKI![]() that renders the mutant enzyme deficient in its ability to phosphorylate the
mPER proteins9.
that renders the mutant enzyme deficient in its ability to phosphorylate the
mPER proteins9.
Casein kinase I delta (CKI![]() )
is highly homologous to CKI
)
is highly homologous to CKI![]() (76% identical at the amino-acid level) and efficiently binds and phosphorylates
mPER1 in vitro10. Our database search
revealed another human casein kinase gene, 'hCKI
(76% identical at the amino-acid level) and efficiently binds and phosphorylates
mPER1 in vitro10. Our database search
revealed another human casein kinase gene, 'hCKI![]() 2'.
This gene is detected in three partially overlapping ESTs that could encode a
carboxy-terminal fragment highly homologous to hCKI
2'.
This gene is detected in three partially overlapping ESTs that could encode a
carboxy-terminal fragment highly homologous to hCKI![]() (91% identical over 73 residues, with two of the substitutions in or near
putative autophosphorylation sites; see Supplementary
Information Figs E and F). Three other novel hCK1 genes were found,
one similar to chicken CKI
(91% identical over 73 residues, with two of the substitutions in or near
putative autophosphorylation sites; see Supplementary
Information Figs E and F). Three other novel hCK1 genes were found,
one similar to chicken CKI![]() 1
(which we name hCKI
1
(which we name hCKI![]() 1),
and two related to hCK1
1),
and two related to hCK1![]() 1,
which we have called hCK1
1,
which we have called hCK1![]() 2
and hCK1
2
and hCK1![]() 3 (see
Supplementary
Information Figs E and F). Functional analysis of CKI
3 (see
Supplementary
Information Figs E and F). Functional analysis of CKI![]() 2
will certainly add to the complexity inherent in the phosphorylation events that
are important for the clockwork.
2
will certainly add to the complexity inherent in the phosphorylation events that
are important for the clockwork.
The timeless mystery
The timeless (tim) gene is essential for circadian function in Drosophila3.
Putative homologues of Drosophila tim have been identified in both
mice and humans (mTim and hTIM, respectively), but placing the
homologue within the murine molecular mechanism has proved difficult2.
Database searches of the completed Drosophila genome show that mammalian
TIM is not the true orthologue of Drosophila TIM, but is the likely
orthologue of a newly described fly gene, timeout (also called tim-2)11,
12. As the core clock functions of TIM in the fly have
been usurped by other clock-relevant genes in the mouse2,
there is no obvious requirement for a mammalian equivalent. Our analysis of
the human genome sequence did not reveal a second Timeless homologue,
consistent with this idea.
Analysis of the completed genome of the worm Caenorhabditis elegans provides additional insight into the timeless mystery. These animals are behavioural outliers, because no circadian rhythms have been reported in the worm. We might reasonably predict that some of the canonical clock genes would be absent from C. elegans. In fact, our searches of the worm databases did not detect any genes with similarity to cryptochromes. Significant hits were obtained for both clock and bmal1 to the same C. elegans gene aha-1 (an hARNT homologue; Table 1), with the similarity far greater for bmal1. The C. elegans gene lin-42, which is required for postembryonic development, shows some similarity in its PAS region to the Drosophila and mammalian PERs13.
Most notably, the worm has only one timeless gene, tim-1, which is highly homologous to Drosophila timeout/tim-2 and mammalian Tim, but substantially removed from the clock-relevant timeless in the fly13. This might suggest that the closely related worm tim-1, mouse/human Tim and fly timeout/tim-2 genes are descendants of an ancestral timeless gene that duplicated in the arthropod lineage after the split with nematodes and vertebrates12. The clock-relevant timeless duplication could then have rapidly diverged to take on a dedicated role in the circadian machinery of insects.
Impact of the human genome on chronobiology
This snapshot analysis of the draft sequence of the human genome provides
candidate sequences that could increase the number of clock genes in mammals.
Aided by further genomic and post-genomic analyses, this could provide new
opportunities for pharmacological manipulation of the human clockwork with 'chronotherapies'
targeted at improving the treatments for those who work shifts or suffer from
jet lag, and those with clock-related sleep or psychiatric problems.
Recently, an autosomal dominant sleep disorder characterized by debilitating
early sleep onset and offset was shown to involve a missense mutation in the CKI![]() binding region of hPER2; the mutation leads to decreased phosphorylation
of hPER2 by CKI
binding region of hPER2; the mutation leads to decreased phosphorylation
of hPER2 by CKI![]() in
vitro14. Linkage analysis identified a
candidate locus in the affected family, and the chromosomal location of known
clock genes from the human genome provided a short-cut in the search. Identified
polymorphisms within human clock genes may aid genetic analysis of other
rhythm-related traits, such as the early riser 'lark' and the late riser 'owl'
phenotypes15.
in
vitro14. Linkage analysis identified a
candidate locus in the affected family, and the chromosomal location of known
clock genes from the human genome provided a short-cut in the search. Identified
polymorphisms within human clock genes may aid genetic analysis of other
rhythm-related traits, such as the early riser 'lark' and the late riser 'owl'
phenotypes15.
The completed human genome sequence will also have a significant impact on the study of circadian output systems. One of the most profound discoveries of the mammalian clock gene 'era' has been the finding that clock genes in mice and humans are widely expressed throughout the body, where they seem to participate in local circadian oscillator function2. These peripheral oscillators are dampened, and require cyclic input from the master clock in the SCN for sustained rhythmicity. Local clock-controlled genes in peripheral organs would provide the flexibility needed to control the circadian rhythms present in these tissues.
Using DNA chip technology, expression profiling of rhythmic gene expression in the SCN and in peripheral tissues will identify candidate output genes and their temporal contribution to local physiology. The completed human genome sequence will then play its part in the identification of these output genes and accelerate fulfilment of one of the great promises of circadian rhythm research: providing a complete understanding of the cellular and molecular events connecting clock genes to circadian behaviour.
Supplementary information is available on Nature's World-Wide Web site (http://www.nature.com) or as paper copy from the London editorial office of Nature.
| 1. | Klein, D. C., Moore, R. Y. & Reppert, S. M. (eds) Suprachiasmatic Nucleus: The Mind's Clock (Oxford Univ. Press, New York, 1991). |
| 2. | Reppert, S. M. & Weaver, D. R. Comparing clockworks: mouse versus fly. J. Biol. Rhythms 15, 357-364 (2000). | PubMed | |
| 3. | Young, M. W. Circadian rhythms. Marking time for a kingdom. Science 288, 451-453 (2000). | Article | PubMed | |
| 4. | Shearman, L. P. et al. Interacting molecular loops in the mammalian circadian clock. Science 288, 1013-1019 (2000). | Article | PubMed | |
| 5. | Dunlap, J. C. Molecular bases for circadian clocks. Cell 96, 271-290 (1999). | PubMed | |
| 6. | Hogenesch, J. B., Gu, Y. Z., Jain, S. & Bradfield, C. A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl Acad. Sci. USA 95, 5474-5479 (1998). | Article | PubMed | |
| 7. | Hogenesch, J. B. et al. The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J. Neurosci. 20, RC83 (2000). | PubMed | |
| 8. | Maemura, K. et al. CLIF, a novel cycle like factor, regulates the circadian oscillation of plasminogen activator inhibitor-1 gene expression. J. Biol. Chem. 275, 36847-36851 (2000). | Article | PubMed | |
| 9. | Lowrey, P. L. et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483-491 (2000). | Article | PubMed | |
| 10. | Vielhaber,
E., Eide, E., Rivers, A., Gao, Z. H. & Virshup, D. M. Nuclear entry
of the circadian regulator mPER1 is controlled by mammalian casein
kinase 1 |
| 11. | Gotter, A. L. et al. A time-less function for mouse Timeless. Nature Neurosci. 3, 755-756 (2000). | Article | PubMed | |
| 12. | Benna, C. et al. A second timeless gene in Drosophila shares greater sequence similarity with mammalian tim. Curr. Biol. 10, R513 (2000). |
| 13. | Jeon, M., Gardner, H. F., Miller, E. A., Deshler, J. & Rougvie, A. E. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286, 1141-1146 (1999). | Article | PubMed | |
| 14. | Toh, K. L. et al. An hPer2 phosphorylation site mutation in familial advanced sleep-phase syndrome. Science, 11 January 2001 (10.1126/science.1057499). | PubMed | |
| 15. | Katzenberg, D. et al. A CLOCK polymorphism associated with human diurnal preference. Sleep 21, 569-576 (1998). | PubMed | |
Acknowledgements. We thank A. Kasprzyk at the European Bioinformatics Institute for assistance. This work was funded by an International Human Frontiers Grant.
Category: 32. Genome Project and Genomics