
Interpretation of microsatellite data
Return to Biology 3466b
Microsatellites are a kind of VNTR (variable number of tandem repeats). In microsatellites, the repeating motifs are usually 2-5 nucleotide long. Different alleles differ purely in the number of repeats. The microsatellite region itself contains short repeating motifs that are found in variable numbers from allele to allele. Microsatellites are amplified by PCR. The primers must target specifically the flanking regions.

Most laboratories visualize microsatellites with a capillary electrophoresis machine similar to a sequencer. However, it is also possible to see them on a polyacrylamide gel, as shown in the following picture, which is reproduced from the Pleurogene website (no longer functional). It is typical for microsatellite bands to exhibit an "echo" when separated on a polyacrylamide gel.
The white grid shows progeny genotypes for 4 different alleles, labelled 1, 2, 3, and 4. The first individual on the left is heterozygous for alleles 1 and 3, etc. The parents, shown in pink and blue on the right, have genotypes 1/4 and 2/3, respectively.
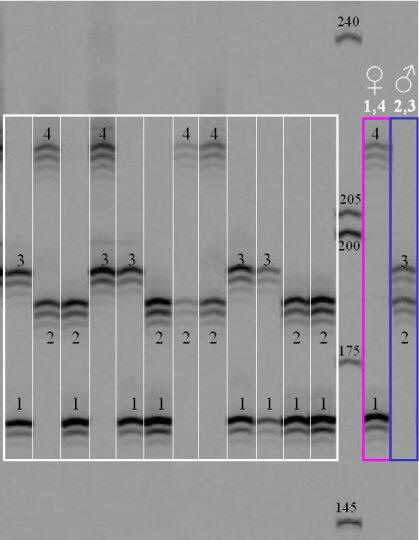
1. How many progeny genotypes are there?
2. Why are neither of the parental genotypes found in the progeny?
Note: The Pleurogene project was a joint Canada-Spain effort to characterize the genetics of turbot.
Return to Biology 3466b